MAIC (Meta-Analysis by Information Content)
Chord diagram showing weighted contributions of each data source about genes implicated in Covid-19 (Nature 2020)
We have developed a new computational approach to do something that is intuitive to biologists, but difficult for computers: to integrate data from diverse sources, weight it according to quality and relevance, and combine it. Our meta-analysis by infomation content (MAIC) algorithm does this, and opens up a range of opportunities across the field of genomics and biology.
You can learn more about the algorithm, and how to use it, at our MAIC project page
-
MAIC fellowship
Development of MAIC was funded in 2013 by a Wellcome-Beit Prize Intermediate Clinical Fellowship (103258/Z/13/Z to K Baillie). This diagram, from the grant application, shows the design for the algorithm. I wrote the code for version 1 of the MAIC package during the first year of the fellowship.
-
MAIC refactoring
In 2018, Andy Law completely refactored the codebase for MAIC, creating a standards-compliant, modular software tool which we shared on the github MAIC repository.
-
MAIC influenza
Nature Communications January 2020
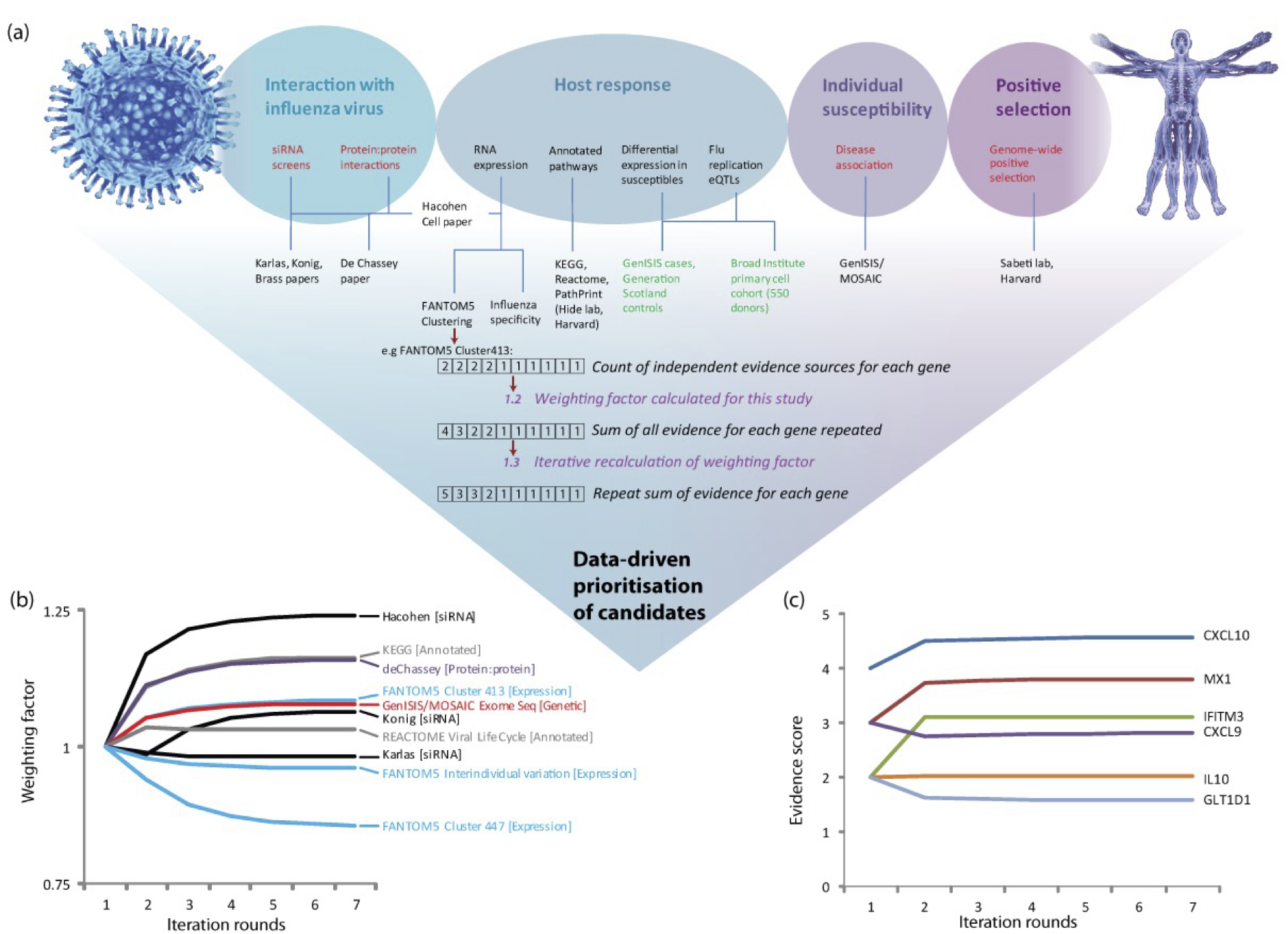
In this paper we reported the design and initial evaluation of the MAIC algorithm, and results of the first genome-wide CRISPR knockout screen of host factors required for replication of influenza virus. The paper was accepted 4 years after I travelled to the Broad Institute of Harvard and MIT to perform a CRISPR screen, together with Bo Li in Nir Hacohen’s lab, because individual validation of the hits was technically challenging. The compeletely new dataset from the CRISPR screen provided a perfect test for the MAIC algorithm. We report a meta-analysis of published work, together with our new screen, in the most comprehensive assessment of host:pathogen interactions with influenza virus to date.
-
MAIC Covid
Scientific Reports December 2020
Throughout the Covid-19 pandemic, the whole lab came together to systematically review the evidence implicating each human protein-coding gene in host:viral interactions and disease pathogenesis.
-
MAIC evaluation
Starting in 2018, Bo Wang performed a systematic comparison of MAIC against other methods for aggregating experimental results. He found that MAIC performs better than other approaches in most circumstances encountered in genomics and biology. The key factor for deciding on an algorithm is the heterogeneity in information quality between input lists. Because MAIC quantifies this heterogeneity, we were able to modify MAIC to tell users if another algorithm will perform better for their datasets.
-
MAIC python package
We released MAIC as a python package which you can install using this command:
pip install pymaic.
Where to next?
Development
We’re working on visualisations of the MAIC approach and on providing a quantitative probability, rather than an arbitrary ranking, for each entity.
Applications
MAIC has a very broad range of potential applications. We’ve used it in the following analyses:
- Influenza: click here to browse MAIC results (Nature Communications 2020)
- Covid-19: click here to browse MAIC results (Scientific Reports 2020, Nature 2021)
Now, we’re working on analyses relating to ARDS, sepsis, and various bacterial genetic screens.