Research themes
-
Apex (altitude physiology expeditions)
In 1999 I started planning to conduct high altitude research in order to better understand the physiology of responses to hypoxia, both in travellers to altitude and in critically ill patients at sea level. Together with several of my university classmates, most notably Roger Thompson, I founded a charity Apex (altitude physiology expeditions), which has organised student-led research expeditions to high altitude every few years ever since.
-
GenISIS grant

My first grant as a chief investigator was for the GenISIS (Genetics of Influenza Susceptibility in Scotland) study.
-
MOSAIC grant

I led the host genetics work for the Wellcome Trust MOSAIC grant (CI: Peter Openshaw), which funded us to extend the GenISIS study by including influenza patients in London and Liverpool.
-
Host genetics review
-
IFITM3
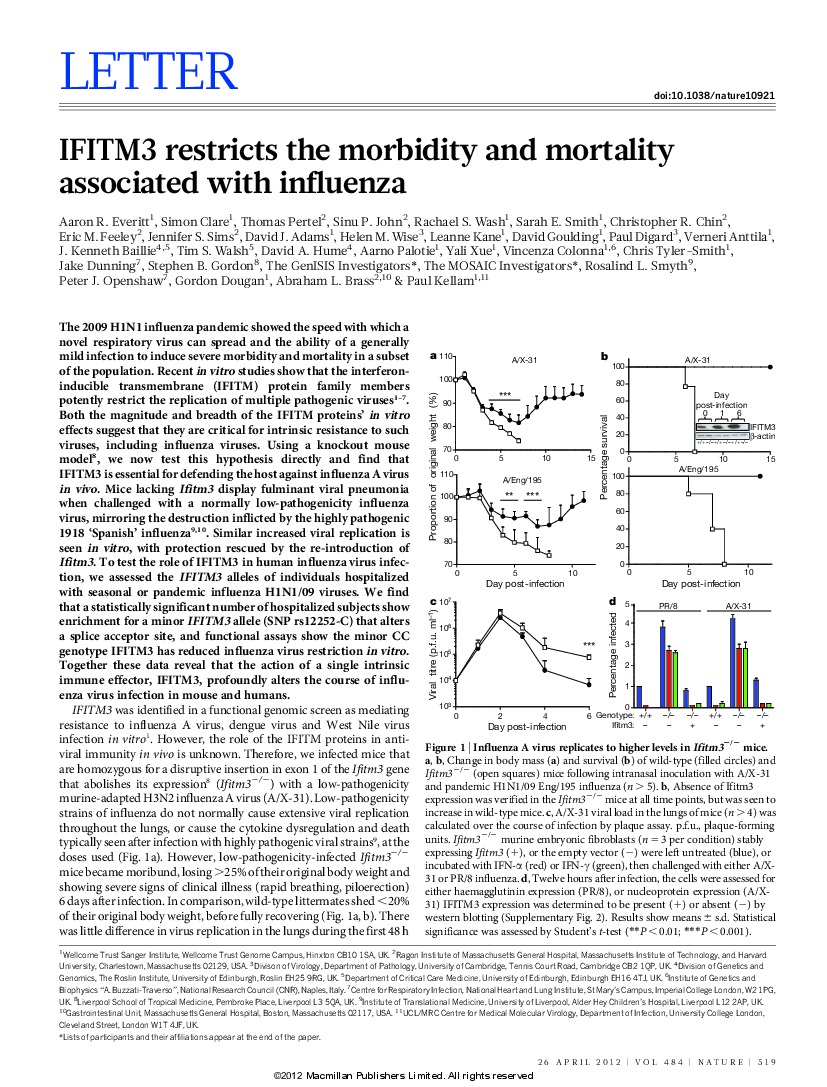
We discovered the first human gene associated with susceptibility to severe influenza
-
Host therapy in influenza
-
MAIC fellowship
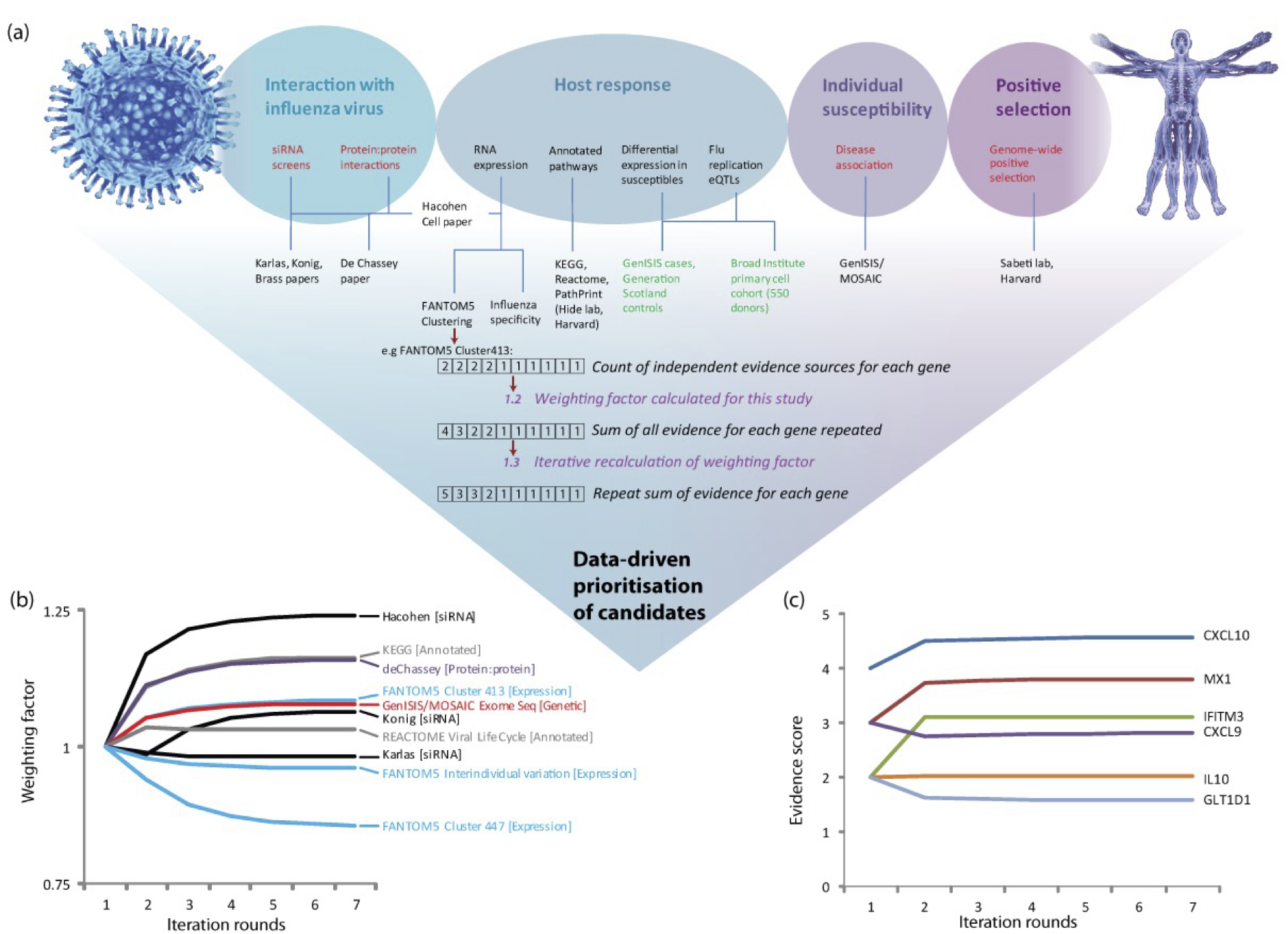
Development of MAIC was funded in 2013 by a Wellcome-Beit Prize Intermediate Clinical Fellowship (103258/Z/13/Z to K Baillie). This diagram, from the grant application, shows the design for the algorithm. I wrote the code for version 1 of the MAIC package during the first year of the fellowship.
-
AMS Clustering

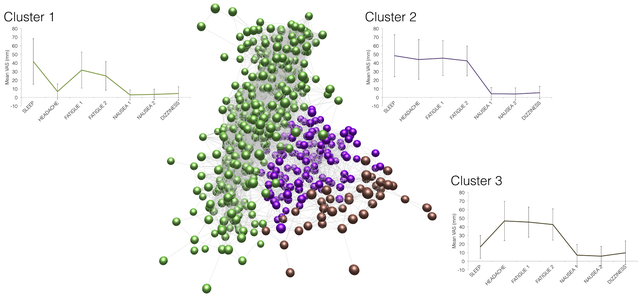
We use a clustering methodology developed for use in transcriptomics, to reveal striking stratification within acute mountain sickness. Our report in arxiv (before biorxiv and medrxiv existed!) showed for the first time that acute mountain sickness is two distinct syndromes that had previously been conflated. This study used data from the first three Apex high altitude research expeditions in 2001, 2003, and 2011.
-
ISARIC CCP
The Lancet Infectious Diseases January 2014
I lead a global consensus process on research response to outbreaks, and write the ISARIC/WHO Clinical Characterisation Protocol.
-
FANTOM5

I demostrate the potential of clustering methodologies to identify genes implicated in biological pathways, including viral response. [@fantom5_2014]
-
Translational genomics

This article describes the fundamental approach that we still take in the lab today: using genetics and functional genomics to find host-directed therapies to promote survival in life-threatening infectious disease.
-
GenOMICC Wellcome Beit grant

The GenOMICC study recieved its first funding from a Wellcome-Beit Prize fellowship.
-
GenOMICC ICS funding

GenOMICC recieved additional funding from the UK Intensive Care Society.
-
Host genetics in viral disease review
Scientific Reports November 2015
We systematically review genes implicated in respiratory viral disease.
-
Treatable traits review
Current Opinion in Systems Biology April 2017

In this review article, we set out the unmet to idenfity treatable traits in critically ill hosts, define relevant terms, and describe systems biology approaches to enable detection of stratification signals in broad clinical syndromes such as sepsis.
-
AMS new consensus
High Altitude Medicine \& Biology March 2018

We redefine acute mountain sickness based on our previous clustering work.
-
Coexpression - a new, independent signal
PLOS Computational Biology March 2018
For more information see baillielab.net/coexpression
-
MAIC refactoring
In 2018, Andy Law completely refactored the codebase for MAIC, creating a standards-compliant, modular software tool which we shared on the github MAIC repository.
-
GenOMICC Sepsis Research FEAT grant

We had a step-change in the GenOMICC when it was supported to expand rapidly by generous commitment from Sepsis Research (Fiona Elizabeth Agnew Trust).
-
Cap snatching
-
Effective shunt
-
Host genetics in influenza review
-
ISARIC CCP Wuhan
Lancet (London, England) January 2020

We didn’t contribute directly to this paper, but it is a fantastic demostration of the effect our decade of work preparing by establishing open source clinical characterisation tool. Our ISARIC CCP was used in this, the first description of novel coronavirus disease in Wuhan.
-
MAIC influenza
Nature Communications January 2020
In this paper we reported the design and initial evaluation of the MAIC algorithm, and results of the first genome-wide CRISPR knockout screen of host factors required for replication of influenza virus. The paper was accepted 4 years after I travelled to the Broad Institute of Harvard and MIT to perform a CRISPR screen, together with Bo Li in Nir Hacohen’s lab, because individual validation of the hits was technically challenging. The compeletely new dataset from the CRISPR screen provided a perfect test for the MAIC algorithm. We report a meta-analysis of published work, together with our new screen, in the most comprehensive assessment of host:pathogen interactions with influenza virus to date.
-
Steroid evidence rapid review
In this short review, we summarise clinical evidence for steroid use in patients with viral lung disease - on balance, suggesting harm overall - and call for trials of steroids in SARS. At the time we thought that although steroids may help some patients, there were likely to harm others. In fact, this is exactly what we subsequently showed in the RECOVERY trial.
-
ISARIC CCP UK Covid
-
Generalisable stratification
We discover that stratification signals are common across different acute illnesses (ARDS and Pancreatitis) For more information see baillielab.net/pancreatitis
-
RECOVERY steroids
New England Journal of Medicine July 2020

We contributed to the setup and delivery of the RECOVERY trial, showing that steroids reduce mortality for a subset of patients with Covid-19.
-
GenOMICC Covid 2020

GenOMICC r1: within 5 months of the first case, we find therapeutic targets, including TYK2, and propose baricitinib. [@pairo-castineirageneticmechanismscritical2021]
-
MAIC Covid
Scientific Reports December 2020
Throughout the Covid-19 pandemic, the whole lab came together to systematically review the evidence implicating each human protein-coding gene in host:viral interactions and disease pathogenesis.
-
GenOMICC WGS funding
Whole Genome Sequencing in GenOMICC was funded in partnership with Genomics England.
-
RECOVERY baricitinib decision
As a direct consequence of the results of GenOMICC, after discussion with SAGE and UK CTAP, baricitinib is included in RECOVERY trial. We chose to add it in addition to steroid treatment and another monoclonal antibody, tocilizumab, which we had already shown to be effective. This was a big risk but, emboldened by the genetic data, we thought it was worth taking.
-
ISARIC Cytokines
-
RECOVERY tocilizumab
We demonstrate in RECOVERY that tocilizumab reduces mortality from severe Covid-19.
-
GenOMICC HGI 2021
PLOS Computational Biology August 2021
We report an additional two variants associated with critical Covid-19, discovered in collaboration with Broad Institute
-
GenOMICC Covid 2021
-
GenOMICC Covid 2021
GenOMICC r3: We discovered multiple new genes associated with critical Covid-19.
-
RECOVERY baricitinib result

In the first-ever direct translation from host genetics to effective drug therapy in infectious disease, we showed in the RECOVERY trial that baricitinib reduces mortality in severe Covid-19.
-
MAIC evaluation
Starting in 2018, Bo Wang performed a systematic comparison of MAIC against other methods for aggregating experimental results. He found that MAIC performs better than other approaches in most circumstances encountered in genomics and biology. The key factor for deciding on an algorithm is the heterogeneity in information quality between input lists. Because MAIC quantifies this heterogeneity, we were able to modify MAIC to tell users if another algorithm will perform better for their datasets.
-
MAIC python package
We released MAIC as a python package which you can install using this command:
pip install pymaic. -
SF94
Nature Communications November 2023
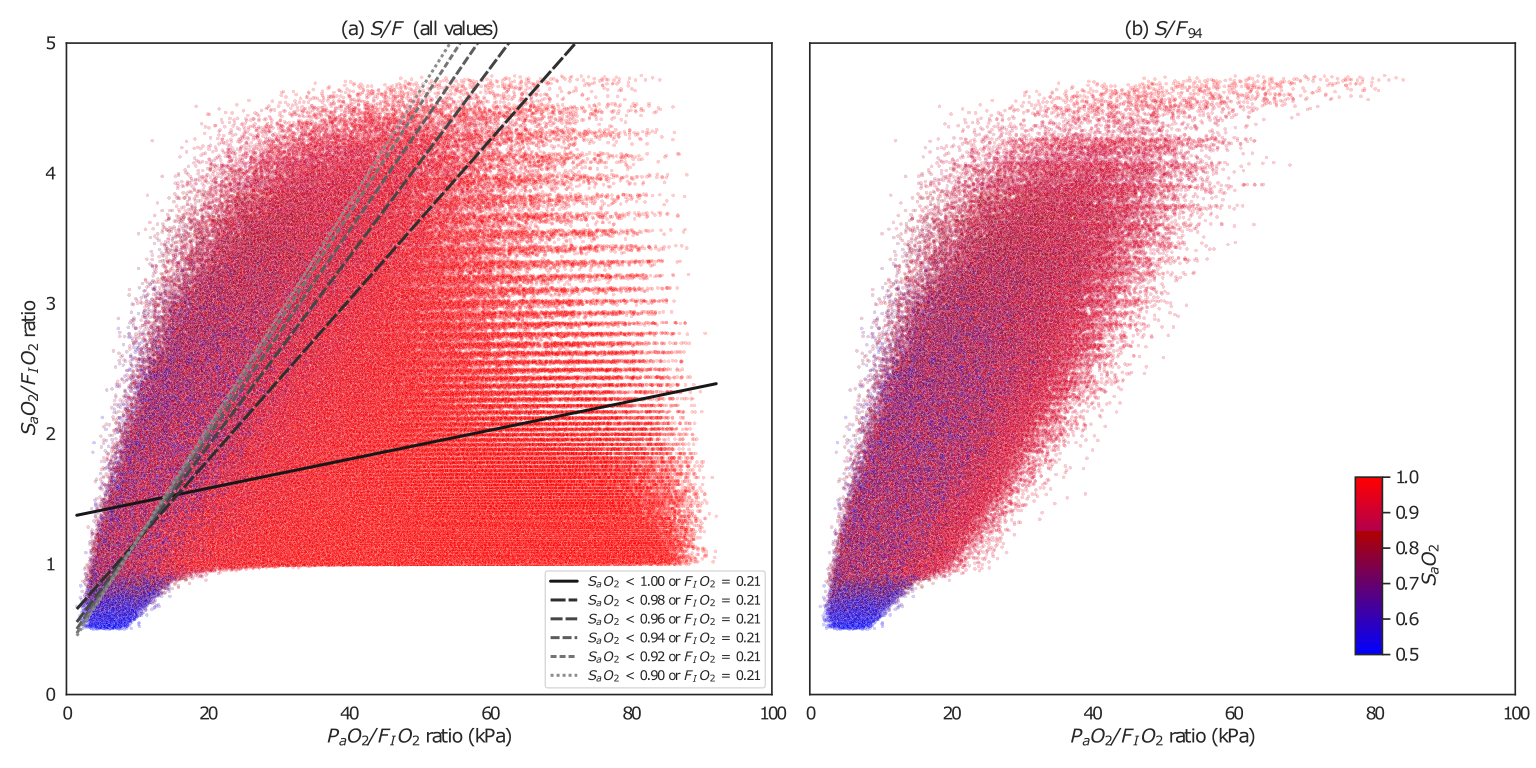
We report a new non-invasive measure of lung oxygenation function (S/F94), evaluate it using physiological modelling data, and quantify the improvement in statistical power from using S/F94 in a clinical trial. We created a completely synthetic dataset that retains the properties of our >300,000 patient ISARIC4C cohort, and built a trial outcomes assessment tool.
-
Altitude physiology model
altitude model
Click here to play with the phsyiological model.
-
ISARIC4C risk score
We created the ISARIC4C score
-
MAIC exploration
David Farr 2018
-
MERS review
MERS review [@arabimiddleeastrespiratory2017]






