Host Genomics

Interpretation of molecular mechanisms explaining genetic associations in Covid-19. Nature, 2023
Host genetics provides the foundation for causal inference in our work. Since 2015 we have run the GenOMICC study to discover new human genetic associations with susceptibility to, and outcome from, critical illness. In 2020, only 5 months after the first Covid-19 patient recruited, we reported a functional genomic analysis in the GenOMICC that suggested a specific drug, baricitinib, would be an effective treatment for critical Covid-19. We went on to show that the treatment was effective in a clinical trial - to our knowledge, the first time this genetics-to-drug-treatment journey has been completed in any infectious disease or critical illness.
-
GenISIS grant

My first grant as a chief investigator was for the GenISIS (Genetics of Influenza Susceptibility in Scotland) study.
-
MOSAIC grant

I led the host genetics work for the Wellcome Trust MOSAIC grant (CI: Peter Openshaw), which funded us to extend the GenISIS study by including influenza patients in London and Liverpool.
-
Host genetics review
-
IFITM3
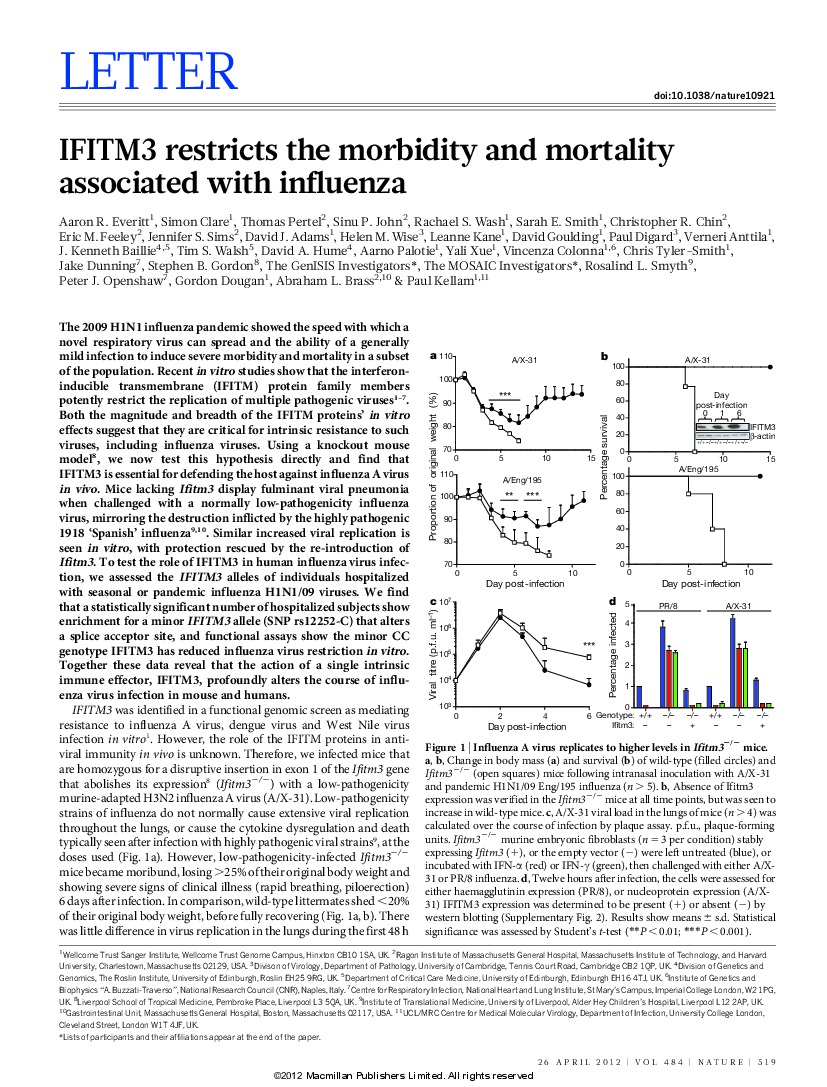
We discovered the first human gene associated with susceptibility to severe influenza
-
FANTOM5

I demostrate the potential of clustering methodologies to identify genes implicated in biological pathways, including viral response. [@fantom5_2014]
-
Translational genomics

This article describes the fundamental approach that we still take in the lab today: using genetics and functional genomics to find host-directed therapies to promote survival in life-threatening infectious disease.
-
GenOMICC Wellcome Beit grant

The GenOMICC study recieved its first funding from a Wellcome-Beit Prize fellowship.
-
GenOMICC ICS funding

GenOMICC recieved additional funding from the UK Intensive Care Society.
-
Host genetics in viral disease review
Scientific Reports November 2015
We systematically review genes implicated in respiratory viral disease.
-
GenOMICC Sepsis Research FEAT grant

We had a step-change in the GenOMICC when it was supported to expand rapidly by generous commitment from Sepsis Research (Fiona Elizabeth Agnew Trust).
-
Host genetics in influenza review
-
GenOMICC Covid 2020

GenOMICC r1: within 5 months of the first case, we find therapeutic targets, including TYK2, and propose baricitinib. [@pairo-castineirageneticmechanismscritical2021]
-
GenOMICC WGS funding
Whole Genome Sequencing in GenOMICC was funded in partnership with Genomics England.
-
RECOVERY baricitinib decision
As a direct consequence of the results of GenOMICC, after discussion with SAGE and UK CTAP, baricitinib is included in RECOVERY trial. We chose to add it in addition to steroid treatment and another monoclonal antibody, tocilizumab, which we had already shown to be effective. This was a big risk but, emboldened by the genetic data, we thought it was worth taking.
-
GenOMICC HGI 2021
PLOS Computational Biology August 2021
We report an additional two variants associated with critical Covid-19, discovered in collaboration with Broad Institute
-
GenOMICC Covid 2021
-
GenOMICC Covid 2021
GenOMICC r3: We discovered multiple new genes associated with critical Covid-19.
-
RECOVERY baricitinib result

In the first-ever direct translation from host genetics to effective drug therapy in infectious disease, we showed in the RECOVERY trial that baricitinib reduces mortality in severe Covid-19.
Where to now?
We are recruiting fast to the GenOMICC study in multiple countries, linking to new data sources, and following up the patients we’ve already recruited. Put simply, we want to do for sepsis, ARDS, influenza, and other forms of critical illness what we have already done for Covid-19: use genetics to find effective treatments.


