Systems biology
Genome-wide unbiased annotation of regulatory elements Nature, 2014
Computational biology is the main focus of the lab. Put simply, we’re trying to explain the molecular mechanisms underlying diseases in patients. We run a large-scale programme to discovery molecular consequences of genetic variation (molQTL). In order to test our predictions, we have a longstanding programme using genome editing in human and porcine cells and tissues to validate potential future drug targets.
-
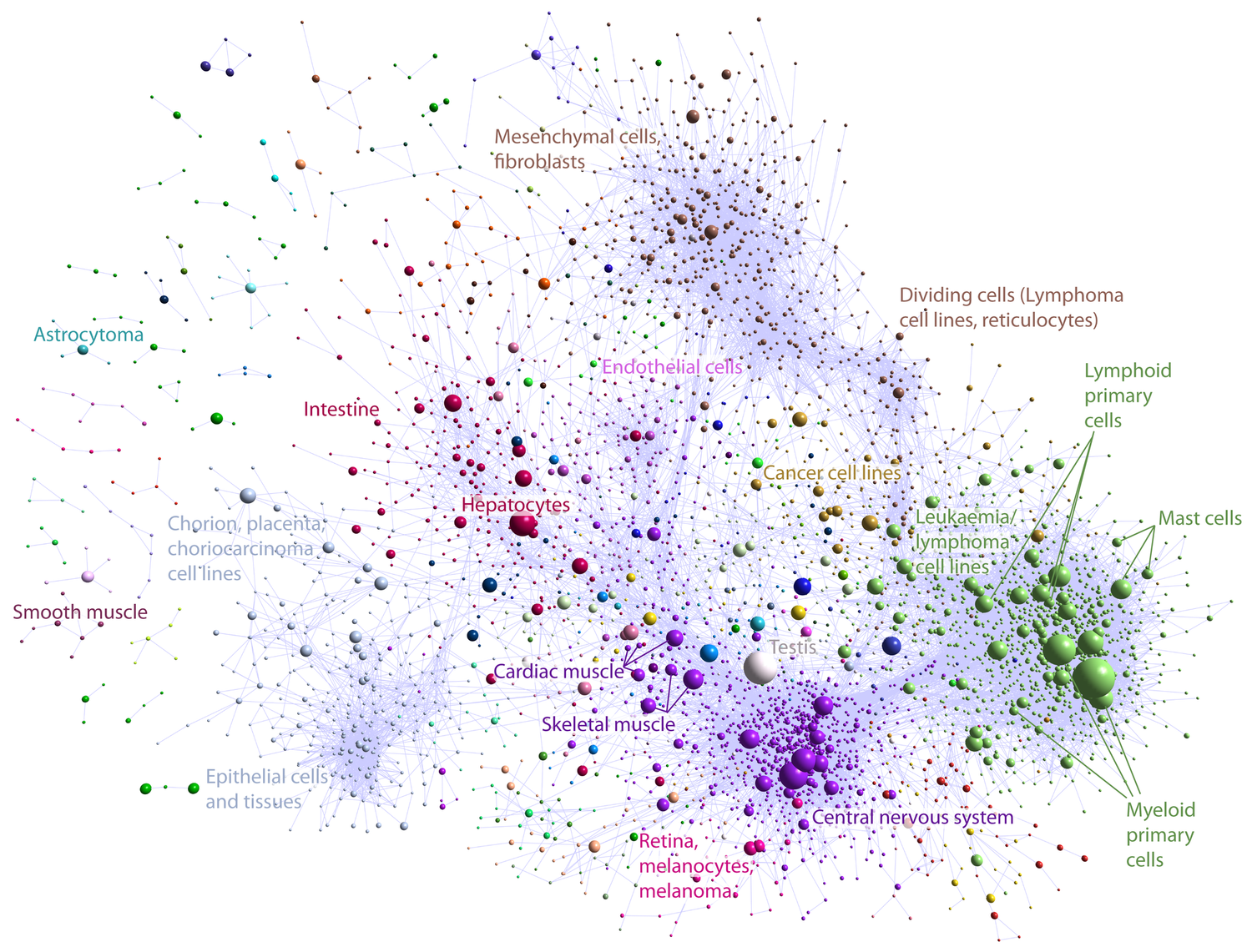
FANTOM5

I demostrate the potential of clustering methodologies to identify genes implicated in biological pathways, including viral response. [@fantom5_2014]
-
Coexpression - a new, independent signal
PLOS Computational Biology March 2018
For more information see baillielab.net/coexpression
-
MAIC influenza
Nature Communications January 2020
In this paper we reported the design and initial evaluation of the MAIC algorithm, and results of the first genome-wide CRISPR knockout screen of host factors required for replication of influenza virus. The paper was accepted 4 years after I travelled to the Broad Institute of Harvard and MIT to perform a CRISPR screen, together with Bo Li in Nir Hacohen’s lab, because individual validation of the hits was technically challenging. The compeletely new dataset from the CRISPR screen provided a perfect test for the MAIC algorithm. We report a meta-analysis of published work, together with our new screen, in the most comprehensive assessment of host:pathogen interactions with influenza virus to date.
Where to now?
We predict that if we study the right cell types and states, then the molecular consequences of disease variants will be detectable. If we can do that, then we can draw causal inferences about disease mechanisms.
